Gas vesicles are hollow structures made of protein found in the cells of certain microorganisms, and researchers believe they can be programmed for use in biomedical applications.
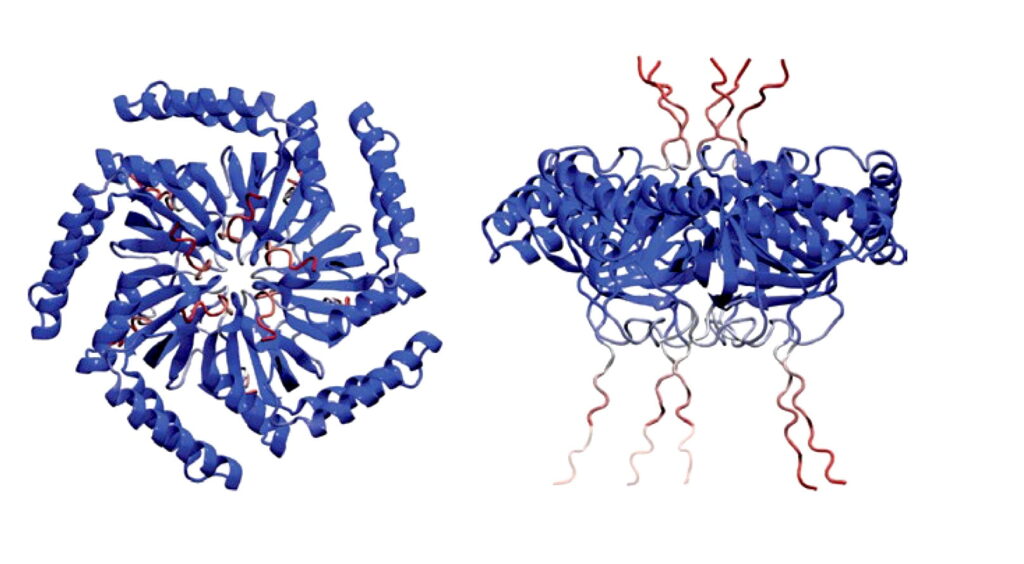
“Inside cells, gas vesicles are packed in a beautiful honeycomb pattern. How this pattern is formed has never been thoroughly understood. We are presenting the first identification of a protein that is responsible for this pattern, and we believe it can be very useful,” said George Lu, an assistant professor of bioengineering and a Cancer Prevention and Research Institute of Texas scholar at Rice University.
Lu and colleagues from Washington University in St. Louis and Duke University have shared their findings in a paper published in Nature Microbiology.
Co-author Yifan Dai, an assistant professor of biomedical engineering at WashU’s McKelvey School of Engineering, said they were drawn to the research with the question of why the vesicles can form in the honeycomb pattern.
With help from WashU colleague Alex Holehouse along with Ashutosh Chilkoti and Lingchong You, of Duke University, the team found this pattern is the most efficient use of space, and the cluster form plays a part in how it functions. Notably, these protein clusters formed in subsaturated solution, a previously identified new form of biological structure, and that drives the organization of these vesicles. Bottom line, they found the function behind this mysterious new form.
